Okay, now we are stepping things up a little bit. This time, the data comes from UCI . According to the website:
For each record in the dataset it is provided: - Triaxial acceleration from the accelerometer (total acceleration) and the estimated body acceleration. - Triaxial Angular velocity from the gyroscope. - A 561-feature vector with time and frequency domain variables. - Its activity label. - An identifier of the subject who carried out the experiment.
The dataset is a bit different - it’s already been divided into train and test sets but they are all text files. Once again, let’s get into it - for fun, let’s also use the same model from last time to see if that architecture still works fine for this dataset. If it doesn’t, let’s see if we can build a different one.
Load Data
The data is split into total, body and gyro - these are the sensor values. There’s also another dataset that has some 561 features extracted from this. Might be fun to play with them but for now, let’s focus on building our CNN network - so this ‘total’ values would be fine for now. May play with .body’ accel values later.
= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/Inertial Signals/total_acc_x_train.txt' # Update this path to your file location # Load into pandas import pandas as pdimport re= pd.read_csv(file_location, sep= ' \ s+' , header= None )
<>:7: SyntaxWarning: invalid escape sequence '\s'
<>:7: SyntaxWarning: invalid escape sequence '\s'
/var/folders/q7/ftl_yg4n1gndbwlnk654hq1h0000gn/T/ipykernel_55236/3576027909.py:7: SyntaxWarning: invalid escape sequence '\s'
data = pd.read_csv(file_location, sep='\s+', header=None)
0
1.012817
1.022833
1.022028
1.017877
1.023680
1.016974
1.017746
1.019263
1.016417
1.020745
...
1.020981
1.018065
1.019638
1.020017
1.018766
1.019815
1.019290
1.018445
1.019372
1.021171
1
1.018851
1.022380
1.020781
1.020218
1.021344
1.020522
1.019790
1.019216
1.018307
1.017996
...
1.019291
1.019258
1.020736
1.020950
1.020491
1.018685
1.015660
1.014788
1.016499
1.017849
2
1.023127
1.021882
1.019178
1.015861
1.012893
1.016451
1.020331
1.020266
1.021759
1.018649
...
1.020304
1.021516
1.019417
1.019312
1.019448
1.019434
1.019916
1.021041
1.022935
1.022019
3
1.017682
1.018149
1.019854
1.019880
1.019121
1.020479
1.020595
1.016340
1.010611
1.009013
...
1.021295
1.022934
1.022183
1.021637
1.020598
1.018887
1.019161
1.019916
1.019602
1.020735
4
1.019952
1.019616
1.020933
1.023061
1.022242
1.020867
1.021939
1.022300
1.022302
1.022254
...
1.022687
1.023670
1.019899
1.017381
1.020389
1.023884
1.021753
1.019425
1.018896
1.016787
5 rows × 128 columns
# Let's see how the labels look like = '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/y_train.txt' # Load = pd.read_csv(file_location, sep= ' \ s+' , header= None )
<>:6: SyntaxWarning: invalid escape sequence '\s'
<>:6: SyntaxWarning: invalid escape sequence '\s'
/var/folders/q7/ftl_yg4n1gndbwlnk654hq1h0000gn/T/ipykernel_55236/3965665900.py:6: SyntaxWarning: invalid escape sequence '\s'
data = pd.read_csv(file_location, sep='\s+', header=None)
Okay looks like there are 7352 segments of windowed data with associated labels
Make a Dataframe
# Load X axis data = '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/Inertial Signals/total_acc_x_train.txt' = pd.read_csv(file_location, sep= ' \ s+' , header= None )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/Inertial Signals/body_acc_x_train.txt' = pd.read_csv(file_loc_body, sep= ' \ s+' , header= None )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/Inertial Signals/body_gyro_x_train.txt' = pd.read_csv(file_loc_gyro, sep= ' \ s+' , header= None )# Append Y axis data = '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/Inertial Signals/total_acc_y_train.txt' = pd.concat([df_total, pd.read_csv(file_location, sep= ' \ s+' , header= None )], axis= 1 )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/Inertial Signals/body_acc_y_train.txt' = pd.concat([df_body, pd.read_csv(file_loc_body, sep= ' \ s+' , header= None )], axis= 1 )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/Inertial Signals/body_gyro_y_train.txt' = pd.concat([df_gyro, pd.read_csv(file_loc_gyro, sep= ' \ s+' , header= None )], axis= 1 )# Z axis = '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/Inertial Signals/total_acc_z_train.txt' = pd.concat([df_total, pd.read_csv(file_location, sep= ' \ s+' , header= None )], axis= 1 )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/Inertial Signals/body_acc_z_train.txt' = pd.concat([df_body, pd.read_csv(file_loc_body, sep= ' \ s+' , header= None )], axis= 1 )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/Inertial Signals/body_gyro_z_train.txt' = pd.concat([df_gyro, pd.read_csv(file_loc_gyro, sep= ' \ s+' , header= None )], axis= 1 )# Labels = '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/train/y_train.txt' = pd.read_csv(file_location, sep= ' \ s+' , header= None )# Subtract 1 from values -= 1
((7352, 384), (7352, 384), (7352, 1))
# Load into 3 dimensional numpy arrays import numpy as np= df_total.values.reshape(7352 , 128 , 3 )= df_body.values.reshape(7352 , 128 , 3 )= df_gyro.values.reshape(7352 , 128 , 3 )= np.array(y_train)
# Do the same for test data # Load X axis data = '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/test/Inertial Signals/total_acc_x_test.txt' = pd.read_csv(file_location, sep= ' \ s+' , header= None )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/test/Inertial Signals/body_acc_x_test.txt' = pd.read_csv(file_loc_body, sep= ' \ s+' , header= None )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/test/Inertial Signals/body_gyro_x_test.txt' = pd.read_csv(file_loc_gyro, sep= ' \ s+' , header= None )# Append Y axis data = '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/test/Inertial Signals/total_acc_y_test.txt' = pd.concat([df_total_test, pd.read_csv(file_location, sep= ' \ s+' , header= None )], axis= 1 )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/test/Inertial Signals/body_acc_y_test.txt' = pd.concat([df_body_test, pd.read_csv(file_loc_body, sep= ' \ s+' , header= None )], axis= 1 )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/test/Inertial Signals/body_gyro_y_test.txt' = pd.concat([df_gyro, pd.read_csv(file_loc_gyro, sep= ' \ s+' , header= None )], axis= 1 )# Z axis = '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/test/Inertial Signals/total_acc_z_test.txt' = pd.concat([df_total_test, pd.read_csv(file_location, sep= ' \ s+' , header= None )], axis= 1 )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/test/Inertial Signals/body_acc_z_test.txt' = pd.concat([df_body_test, pd.read_csv(file_loc_body, sep= ' \ s+' , header= None )], axis= 1 )= '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/test/Inertial Signals/body_gyro_z_test.txt' = pd.concat([df_gyro, pd.read_csv(file_loc_gyro, sep= ' \ s+' , header= None )], axis= 1 )# Labels = '/Users/manikandanravi/Code/quarto-test/uci-har-dataset/test/y_test.txt' = pd.read_csv(file_location, sep= ' \ s+' , header= None )-= 1 # Convert to numpy arrays = df_total_test.values.reshape(len (df_total_test), 128 , 3 )= df_body_test.values.reshape(len (df_body_test), 128 , 3 )= df_gyro.values.reshape(len (df_gyro), 128 , 3 )= np.array(y_test)print ("Shapes of test data:" )print ("X_total_test:" , X_total_test.shape)print ("X_body_test:" , X_body_test.shape)print ("X_gyro_test:" , X_gyro_test.shape)print ("y_test:" , y_test.shape)
OKay, now we have the data. Let’s get to work on building dataloaders and the models
Build Model
Data Loader
import torchimport torch.nn as nnfrom torch.utils.data import Dataset, DataLoader# Dataset wrapper class UCIDatasetWrapper(Dataset):def __init__ (self , X, y):# Convert (N, T, C) -> (N, C, T) for Conv1D self .X = torch.tensor(X.transpose(0 ,2 ,1 ), dtype= torch.float32)self .y = torch.tensor(y, dtype= torch.long )def __len__ (self ):return len (self .y)def __getitem__ (self , i):return self .X[i], self .y[i]
= UCIDatasetWrapper(X_total_train, y_train)= UCIDatasetWrapper(X_body_train, y_train)= UCIDatasetWrapper(X_total_test, y_test)= UCIDatasetWrapper(X_body_test, y_test)= DataLoader(train_ds, batch_size= 32 , shuffle= True )= DataLoader(train_ds_body, batch_size= 32 , shuffle= True )= DataLoader(test_ds, batch_size= 32 , shuffle= False )
Model
Lets use the same model from the previous notbook
# 1D CNN model (Same as our previous notebook) import torch.nn.functional as Fclass HAR1DCNN(nn.Module):def __init__ (self , in_channels= 3 , n_classes= 6 ): # adjust n_classes! super ().__init__ ()self .conv1 = nn.Conv1d(in_channels, 32 , kernel_size= 3 , padding= 1 )self .bn1 = nn.BatchNorm1d(32 )self .conv2 = nn.Conv1d(32 , 64 , kernel_size= 3 , padding= 1 )self .bn2 = nn.BatchNorm1d(64 )self .pool = nn.MaxPool1d(2 )self .conv3 = nn.Conv1d(64 , 128 , kernel_size= 3 , padding= 1 )self .bn3 = nn.BatchNorm1d(128 )self .global_pool = nn.AdaptiveAvgPool1d(1 )self .fc = nn.Linear(128 , n_classes)def forward(self , x):= F.relu(self .bn1(self .conv1(x)))= self .pool(x) # (N, 32, 20) = F.relu(self .bn2(self .conv2(x)))= self .pool(x) # (N, 64, 10) = F.relu(self .bn3(self .conv3(x)))= self .global_pool(x).squeeze(- 1 ) # (N, 128) return self .fc(x)
Train the model
= torch.device("cuda" if torch.cuda.is_available() else "cpu" )if device.type != "cuda" := torch.device("mps" if torch.backends.mps.is_available() else "cpu" )= len (torch.unique(torch.tensor(y_train)))= HAR1DCNN(in_channels= 3 , n_classes= n_classes).to(device)
# Optimizer and loss function = torch.optim.Adam(model.parameters(), lr= 1e-3 )= nn.CrossEntropyLoss()
def run_epoch(loader, train= True ):if train:else :eval ()= 0 , 0 , 0 for Xb, yb in loader:= Xb.to(device), yb.to(device)if train:= model(Xb)= loss_fn(out, yb.squeeze()) # Removed - 1 if train:+= loss.item() * Xb.size(0 )= out.argmax(1 )+= (preds == yb.squeeze()).sum ().item() # Removed - 1 += Xb.size(0 )return total_loss/ total, correct/ total
for epoch in range (10 ):= run_epoch(train_dl)= run_epoch(test_dl, train= False )print (f'Epoch { epoch+ 1 } : train_loss= { train_loss:.4f} , train_acc= { train_acc:.4f} , test_loss= { test_loss:.4f} , test_acc= { test_acc:.4f} ' )
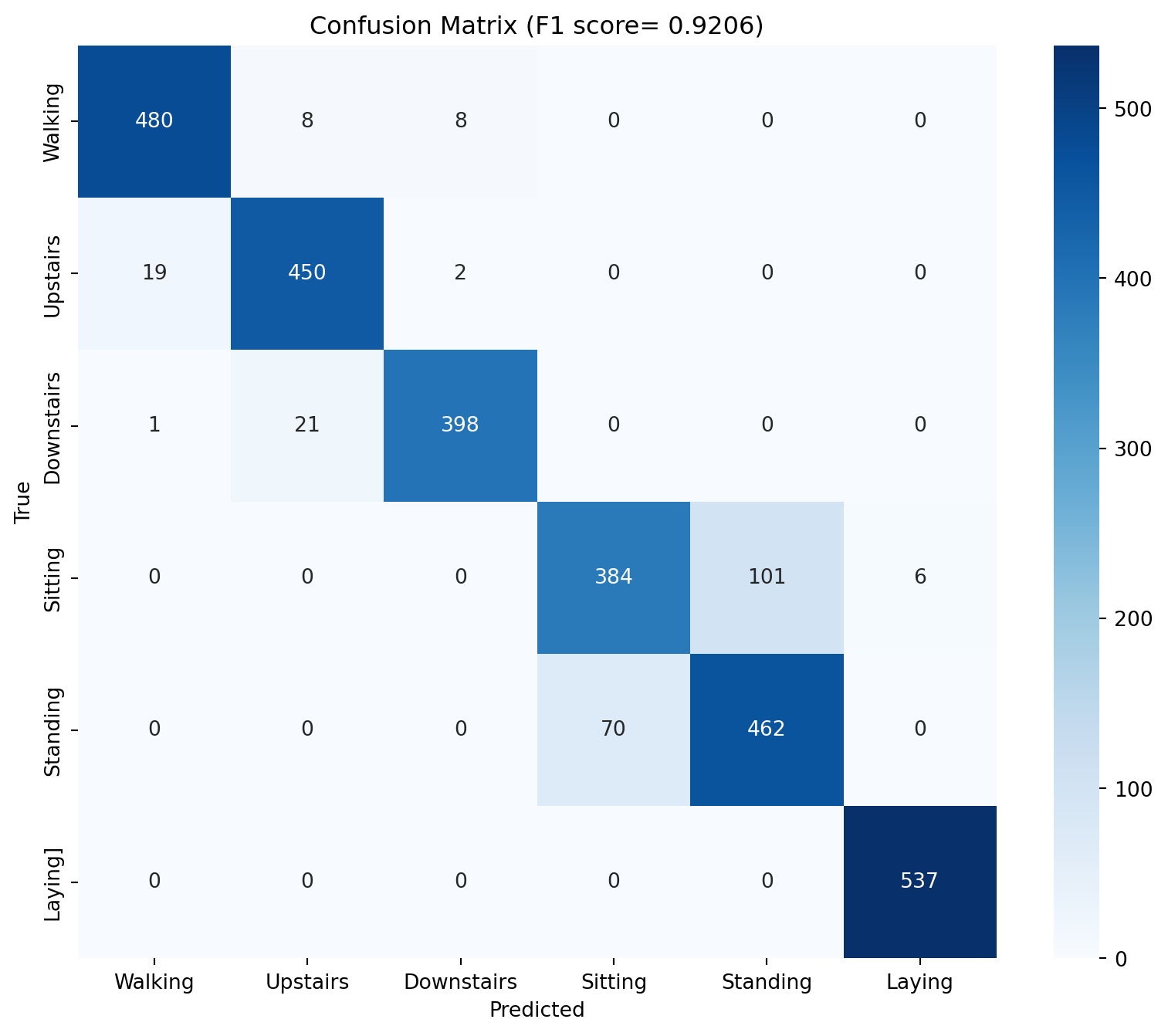
Epoch 1: train_loss=0.5877, train_acc=0.8232, test_loss=0.3945, test_acc=0.8354
Epoch 2: train_loss=0.2595, train_acc=0.9083, test_loss=0.3221, test_acc=0.8856
Epoch 3: train_loss=0.2047, train_acc=0.9210, test_loss=0.2862, test_acc=0.8873
Epoch 4: train_loss=0.1797, train_acc=0.9320, test_loss=0.2683, test_acc=0.8761
Epoch 5: train_loss=0.1613, train_acc=0.9357, test_loss=0.3262, test_acc=0.8649
Epoch 6: train_loss=0.1549, train_acc=0.9377, test_loss=0.2309, test_acc=0.9087
Epoch 7: train_loss=0.1429, train_acc=0.9407, test_loss=0.2487, test_acc=0.9097
Epoch 8: train_loss=0.1482, train_acc=0.9418, test_loss=0.2335, test_acc=0.9169
Epoch 9: train_loss=0.1367, train_acc=0.9450, test_loss=0.2720, test_acc=0.9040
Epoch 10: train_loss=0.1353, train_acc=0.9452, test_loss=0.2988, test_acc=0.8843
Evaluate model
# Evaluate the model - get F1 score, confusion matrix from sklearn.metrics import f1_score, confusion_matrixeval ()= []= []with torch.no_grad():for Xb, yb in test_dl:= Xb.to(device), yb.to(device)= model(Xb)= out.argmax(1 )= f1_score(y_true, y_pred, average= 'macro' )= confusion_matrix(y_true, y_pred)print (f'F1 score: { f1:.4f} ' )# PLot confusion matrix import seaborn as snsimport matplotlib.pyplot as plt= (10 , 8 ))= True , fmt= 'd' , cmap= 'Blues' )'Predicted' )'True' )f'Confusion Matrix (F1 score= { f1:.4f} )' )0.5 , 1.5 , 2.5 , 3.5 , 4.5 , 5.5 ], ['Walking' , 'Upstairs' , 'Downstairs' , 'Sitting' , 'Standing' , 'Laying' ])0.5 , 1.5 , 2.5 , 3.5 , 4.5 , 5.5 ], ['Walking' , 'Upstairs' , 'Downstairs' , 'Sitting' , 'Standing' , 'Laying]' ])
Changing Data
What if we train the model just with the ‘body’ accelerometer data?
Just to recap: > The sensor signals (accelerometer and gyroscope) were pre-processed by applying noise filters and then sampled in fixed-width sliding windows of 2.56 sec and 50% overlap (128 readings/window). The sensor acceleration signal, which has gravitational and body motion components, was separated using a Butterworth low-pass filter into body acceleration and gravity. The gravitational force is assumed to have only low frequency components, therefore a filter with 0.3 Hz cutoff frequency was used. From each window, a vector of features was obtained by calculating variables from the time and frequency domain.
for epoch in range (10 ):= run_epoch(train_dl_body)= run_epoch(test_dl, train= False )print (f'Epoch { epoch+ 1 } : train_loss= { train_loss:.4f} , train_acc= { train_acc:.4f} , test_loss= { test_loss:.4f} , test_acc= { test_acc:.4f} ' )
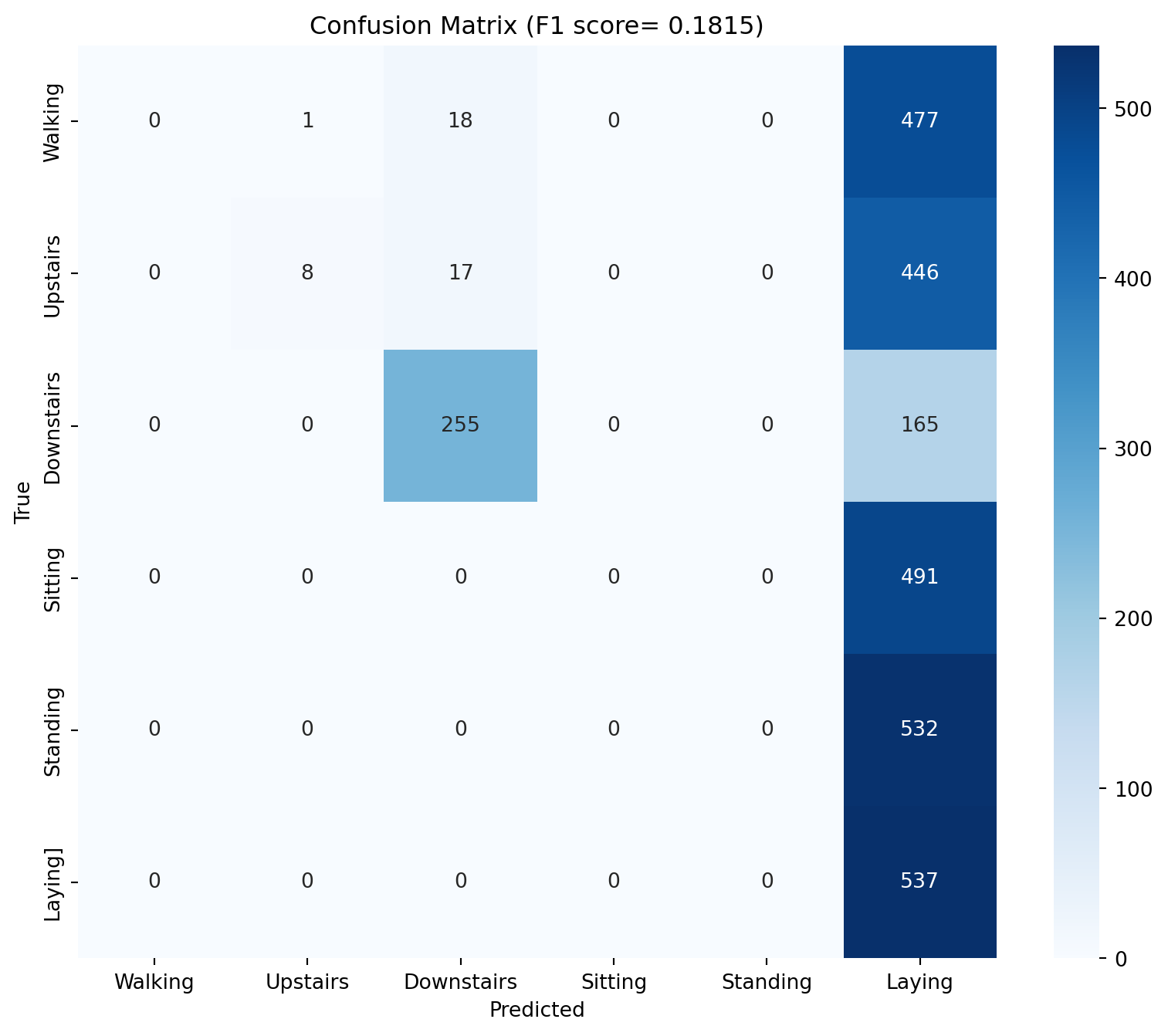
Epoch 1: train_loss=0.7459, train_acc=0.6017, test_loss=5.3111, test_acc=0.4411
Epoch 2: train_loss=0.6667, train_acc=0.6245, test_loss=5.0896, test_acc=0.3006
Epoch 3: train_loss=0.6550, train_acc=0.6304, test_loss=7.9090, test_acc=0.2694
Epoch 4: train_loss=0.6306, train_acc=0.6383, test_loss=9.0907, test_acc=0.1968
Epoch 5: train_loss=0.6254, train_acc=0.6510, test_loss=10.1547, test_acc=0.2158
Epoch 6: train_loss=0.6231, train_acc=0.6508, test_loss=10.2332, test_acc=0.2063
Epoch 7: train_loss=0.6294, train_acc=0.6502, test_loss=12.2881, test_acc=0.2172
Epoch 8: train_loss=0.6161, train_acc=0.6564, test_loss=16.7289, test_acc=0.1849
Epoch 9: train_loss=0.6094, train_acc=0.6585, test_loss=16.9381, test_acc=0.1822
Epoch 10: train_loss=0.6113, train_acc=0.6586, test_loss=18.3700, test_acc=0.1822
# Evaluate the model - get F1 score, confusion matrix from sklearn.metrics import f1_score, confusion_matrixeval ()= []= []with torch.no_grad():for Xb, yb in test_dl:= Xb.to(device), yb.to(device)= model(Xb)= out.argmax(1 )= f1_score(y_true, y_pred, average= 'macro' )= confusion_matrix(y_true, y_pred)print (f'F1 score: { f1:.4f} ' )# PLot confusion matrix import seaborn as snsimport matplotlib.pyplot as plt= (10 , 8 ))= True , fmt= 'd' , cmap= 'Blues' )'Predicted' )'True' )f'Confusion Matrix (F1 score= { f1:.4f} )' )0.5 , 1.5 , 2.5 , 3.5 , 4.5 , 5.5 ], ['Walking' , 'Upstairs' , 'Downstairs' , 'Sitting' , 'Standing' , 'Laying' ])0.5 , 1.5 , 2.5 , 3.5 , 4.5 , 5.5 ], ['Walking' , 'Upstairs' , 'Downstairs' , 'Sitting' , 'Standing' , 'Laying]' ])
As we can see (and intutitively expect), if we take out the gravitational component, the accuracy values are pretty poor. What if we feed all these 9 vectors in?
Running the model with all the data
# Concatenate total, body and gyro = np.concat((X_total_train, X_body_train, X_gyro_train), axis= 2 )# We should have 9 channels now = np.concat((X_total_test, X_body_test, X_gyro_test), axis= 2 )
# Create data loaders = UCIDatasetWrapper(X_all_train, y_train)= DataLoader(train_ds_all, batch_size= 32 , shuffle= True )= UCIDatasetWrapper(X_all_test, y_test)= DataLoader(test_ds_all, batch_size= 32 , shuffle= False )
# We need a new model with 9 channels class HAR1DCNN_9ch(nn.Module):def __init__ (self , in_channels= 9 , n_classes= 6 ): # adjust n_classes! super ().__init__ ()self .conv1 = nn.Conv1d(in_channels, 32 , kernel_size= 3 , padding= 1 )self .bn1 = nn.BatchNorm1d(32 )self .conv2 = nn.Conv1d(32 , 64 , kernel_size= 3 , padding= 1 )self .bn2 = nn.BatchNorm1d(64 )self .pool = nn.MaxPool1d(2 )self .conv3 = nn.Conv1d(64 , 128 , kernel_size= 3 , padding= 1 )self .bn3 = nn.BatchNorm1d(128 )self .global_pool = nn.AdaptiveAvgPool1d(1 )self .fc = nn.Linear(128 , n_classes)def forward(self , x):= F.relu(self .bn1(self .conv1(x)))= self .pool(x) # (N, 32, 20) = F.relu(self .bn2(self .conv2(x)))= self .pool(x) # (N, 64, 10) = F.relu(self .bn3(self .conv3(x)))= self .global_pool(x).squeeze(- 1 ) # (N, 128) return self .fc(x)
Train the new model
= torch.device("cuda" if torch.cuda.is_available() else "cpu" )if device.type != "cuda" := torch.device("mps" if torch.backends.mps.is_available() else "cpu" )= len (torch.unique(torch.tensor(y_train)))= HAR1DCNN_9ch(in_channels= 9 , n_classes= n_classes).to(device)
# Optimizer and loss function = torch.optim.Adam(model_2.parameters(), lr= 1e-3 )= nn.CrossEntropyLoss()
def run_epoch(loader, train= True ):if train:else :eval ()= 0 , 0 , 0 for Xb, yb in loader:= Xb.to(device), yb.to(device)if train:= model_2(Xb)= loss_fn(out, yb.squeeze())if train:+= loss.item() * Xb.size(0 )= out.argmax(1 )+= (preds == yb.squeeze()).sum ().item()+= Xb.size(0 )return total_loss/ total, correct/ total
for epoch in range (20 ):= run_epoch(train_dl_all)= run_epoch(test_dl_all, train= False )print (f'Epoch { epoch+ 1 } : train_loss= { train_loss:.4f} , train_acc= { train_acc:.4f} , test_loss= { test_loss:.4f} , test_acc= { test_acc:.4f} ' )
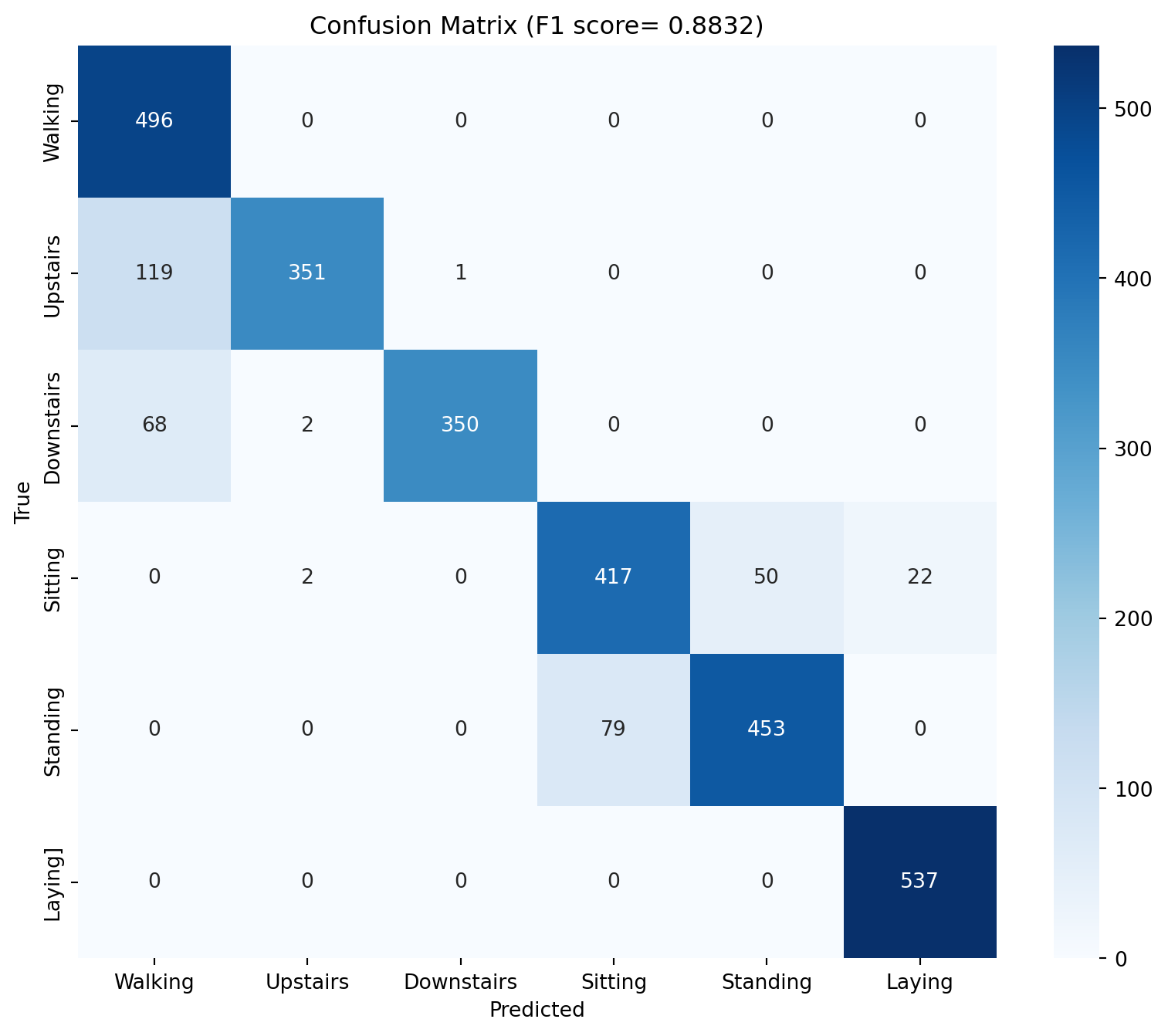
Epoch 1: train_loss=0.5696, train_acc=0.8290, test_loss=0.3161, test_acc=0.8901
Epoch 2: train_loss=0.2113, train_acc=0.9245, test_loss=0.2400, test_acc=0.9111
Epoch 3: train_loss=0.1720, train_acc=0.9323, test_loss=0.2616, test_acc=0.9050
Epoch 4: train_loss=0.1541, train_acc=0.9378, test_loss=0.5100, test_acc=0.8168
Epoch 5: train_loss=0.1456, train_acc=0.9404, test_loss=0.2445, test_acc=0.9175
Epoch 6: train_loss=0.1269, train_acc=0.9461, test_loss=0.2456, test_acc=0.9192
Epoch 7: train_loss=0.1234, train_acc=0.9468, test_loss=0.2176, test_acc=0.9298
Epoch 8: train_loss=0.1274, train_acc=0.9463, test_loss=0.2412, test_acc=0.9057
Epoch 9: train_loss=0.1156, train_acc=0.9504, test_loss=0.2075, test_acc=0.9335
Epoch 10: train_loss=0.1155, train_acc=0.9506, test_loss=0.2143, test_acc=0.9325
Epoch 11: train_loss=0.1161, train_acc=0.9506, test_loss=0.2402, test_acc=0.9247
Epoch 12: train_loss=0.1132, train_acc=0.9510, test_loss=0.2097, test_acc=0.9318
Epoch 13: train_loss=0.1122, train_acc=0.9504, test_loss=0.2070, test_acc=0.9416
Epoch 14: train_loss=0.1088, train_acc=0.9551, test_loss=0.2278, test_acc=0.9386
Epoch 15: train_loss=0.1059, train_acc=0.9561, test_loss=0.2006, test_acc=0.9430
Epoch 16: train_loss=0.1069, train_acc=0.9547, test_loss=0.3617, test_acc=0.8761
Epoch 17: train_loss=0.1043, train_acc=0.9559, test_loss=0.2004, test_acc=0.9460
Epoch 18: train_loss=0.1017, train_acc=0.9570, test_loss=0.2802, test_acc=0.9138
Epoch 19: train_loss=0.1041, train_acc=0.9566, test_loss=0.2089, test_acc=0.9437
Epoch 20: train_loss=0.1043, train_acc=0.9539, test_loss=0.2495, test_acc=0.9145
# Evaluate the model - get F1 score, confusion matrix from sklearn.metrics import f1_score, confusion_matrixeval ()= []= []with torch.no_grad():for Xb, yb in test_dl_all:= Xb.to(device), yb.to(device)= model_2(Xb)= out.argmax(1 )= f1_score(y_true, y_pred, average= 'macro' )= confusion_matrix(y_true, y_pred)print (f'F1 score: { f1:.4f} ' )# PLot confusion matrix import seaborn as snsimport matplotlib.pyplot as plt= (10 , 8 ))= True , fmt= 'd' , cmap= 'Blues' )'Predicted' )'True' )f'Confusion Matrix (F1 score= { f1:.4f} )' )0.5 , 1.5 , 2.5 , 3.5 , 4.5 , 5.5 ], ['Walking' , 'Upstairs' , 'Downstairs' , 'Sitting' , 'Standing' , 'Laying' ])0.5 , 1.5 , 2.5 , 3.5 , 4.5 , 5.5 ], ['Walking' , 'Upstairs' , 'Downstairs' , 'Sitting' , 'Standing' , 'Laying]' ])
As we can see, this has improved on the accuracy on our first model (albeit, not by a lot - because our first model was already quite good).